The early approaches used by Forest Research involving biochemical markers such as terpenes and isozymes have been largely superseded by DNA based methods.
Polymerase chain reaction (PCR)
The DNA based methods utilise the polymerase chain reaction (PCR) to copy and multiply up small regions of the genome using short nucleotide sequences known as primers. The initial PCR methods that were used were RAPDs (Random Amplified Polymorphic DNA) and these required no prior knowledge of the genome in order to work.
Microsatellites
We have now progressed to more sophisticated approaches such as microsatellites which require sequence information in order to work. Microsatellites are regions of short tandem repeats which tend to be highly variable. This feature makes them very useful for genotyping individuals and exploring the genetic structure of populations and the gene flow occurring within them.
Species studied
The molecular laboratory at Forest Research has worked on the following species:
Real-time PCR
As an approach to monitoring black grouse populations, we developed a real-time PCR based assay to distinguish between faecal droppings from black grouse and two other tetraonid species that can occur sympatrically; red grouse (Lagopus lagopus scotica) and capercaillie (Tetrao urogallus) in order to monitor black grouse populations.
The real-time PCR approach is several orders of magnitude more sensitive than traditional PCR methods and is, therefore, appropriate for field collected faecal samples which tend to yield poor quality DNA in low copy number.
Completed EU funded genetic conservation studies
This page gives information on the following completed EU-funded genetic conservation studies:
- FAIROAK
- OAKFLOW
- EUROPOP
- FRAXIGEN
- GENOSILVA
- EUFGIS
Synthetic maps of gene diversity and provenance performance for utilisation and conservation of oak resources in Europe (FAIROAK)
Completed in 2000
Oak species represent a major component of the European forest resource. They supply quality wood, stabilise forests and enhance biological richness in forest eco-systems. Despite the silvicultural and economic importance of oaks, the knowledge of their genetic diversity is poorly understood. This hampers genetic improvement programmes, decisions about seed transfers, and choice of provenances from plantations. The more the species is used to afforest agricultural lands and to enrich existing forest currently in monoculture, the more information about the genetic diversity will become important.
The aim of this project was to provide geneticists, ecologists and foresters with an integrated description of oak (Quercus petraea and Q. robur) genetic resources in the form of synthetic maps based on chloroplast DNA (cpDNA) polymorphism and provenance variation.
Forest Research was involved with the following activities:
- Building a geographic map of cpDNA haplotypes for GB
- Analysing provenance tests on a range wide scale so that inferences may be drawn for seed transfer rules, comparing the provenance results with the cpDNA results in order to verify if there is any relationship between the geographic variation of growth and adaptive traits and the maternal lineage of the population
- Evaluating the level of diversity in the Q. petraea and Q. robur species and its geographic variation, by sampling large size populations and using hypervariable markers (exhibiting numerous alleles).
FAIROAK website
Intra and interspecific gene flow in oaks (OAKFLOW)
Completed December 2004
Examined intra and interspecific gene flows in oaks, as mechanisms promoting genetic diversity and adaptive potential. Forest services and conservation agencies were closely involved in testing various implications of gene flow in management and conservation issues.
Forest Research involvement in OAKFLOW project
OAKFLOW website
Genetic diversity in the river populations of the European black poplar (EUROPOP)
Completed in 2000
The European black poplar (Populus nigra) is a unique pioneer species belonging to the riparian ecosystems. It has social and economic importance and is an ideal species for the study of conservation biology.
The aim of this project was to develop strategies for the conservation of European black poplar and its restoration in riparian ecosystems, based on the measurement of the genetic diversity in wild populations at different levels.
Forest Research was involved with the following activities:
- Standardisation of methods
- Describing the genetic diversity and studying the scale of gene exchange within and between river systems
- Verification of existing gene banks
- Studying key parameters of stand dynamics for in situ management and re-introduction strategies
- Developing strategies for the conservation of P. nigra and its restoration to riparian ecosystems based on the measurement of the genetic diversity in wild populations
- Describing the genetic diversity in ex situ collections in order to evaluate the current state of conservation in Europe
- Formulating guidelines for in situ management and re-introduction strategies.
Ash trees for the future (FRAXIGEN)
Complete in June 2005
The scientific objectives of FRAXIGEN were to:
- Study patterns of gene flow and genetic diversity in three European Fraxinus species, and how these are influenced by variation in reproductive systems
- Study how natural ash populations have adapted to their environment, and how anthropogenic selection for productive characters has affected adaptive variation
- Provide guidance for governmental, private and public interest groups on the collection, exploitation, and conservation of ash genetic resources.
FRAXIGEN website
European forest genomics network (GENOSILVA)
COST Action E28 – Completed February 2006
Project objectives:
- Use genome resource developed for model tree species linking gene sequence and gene function to enhance understanding of the genetic and cellular processes affecting tree growth and survival.
- Use understanding to develop new tools to enhance forest productivity Dialogue with forestry practitioners, tree breeders, forest owners, forest managers and policymakers.
- Inform the public of the use of new genetic tools and technologies; to show benefits.
GENOSILVA EU web page
Establishment of a European information system on forest genetic resources (EUFGIS)
Ongoing, due to end September 2010
EUFGIS aims at establishing a Web-based information system to serve as a documentation platform for national FGR inventories and to support practical implementation of gene conservation and sustainable forest management in Europe. The project will create a network of national focal persons in European countries to provide updated data for the information system once it has been established.
Before the information system can be developed, the major effort is to harmonize minimum requirements for dynamic gene conservation units of forest trees and develop common information standards for these units at pan-European level.
The project will also provide training on FGR documentation to national focal persons.
EUFGIS website
Related pages
Genetic conservation molecular studies of black poplar
 Black poplar is Britain’s rarest native tree and it is estimated that only about 7000 individual trees remain of this once common tree in Britain. It has become endangered because its natural habitat of flooded winter meadows has been largely lost due to drainage of agricultural land, canalisation of rivers and urbanisation. As a result of this the British population is unable to regenerate sexually and many of the existing trees are ancient and nearing the end of their natural lives. There has been little vegetative propagation of the species in Britain since the introduction of the faster growing P. x euramericana hybrids in the nineteenth century.
Black poplar is Britain’s rarest native tree and it is estimated that only about 7000 individual trees remain of this once common tree in Britain. It has become endangered because its natural habitat of flooded winter meadows has been largely lost due to drainage of agricultural land, canalisation of rivers and urbanisation. As a result of this the British population is unable to regenerate sexually and many of the existing trees are ancient and nearing the end of their natural lives. There has been little vegetative propagation of the species in Britain since the introduction of the faster growing P. x euramericana hybrids in the nineteenth century.
Clonal duplication
Prior to our studies it was known that the species had been extensively propagated by cuttings, but it was not known how many individual genotypes were represented in the British population and what the level of clonal duplication was.
Our early work on this species was done using RAPDs – Random Amplified Polymorphic DNA – (Cottrell, Forrest & White, 1997) and the results were later confirmed using microsatellites in the framework of the EU funded project, EUROPOP (Storme et al., 2004). In a sample of 88 British trees only 20 individual clones were discovered, confirming suspicions that the British population is highly clonal.
Glacial origins
Chloroplast DNA (cpDNA) is a slowly evolving, haploid genome which is maternally inherited. It offers an ideal method of studying post glacial colonisation routes.
Forest Research collaborated with other laboratories in the EUROPOP project to determine the routes taken by black poplar when it recolonised Europe following the end of the last ice age. These studies clearly showed that there were at least two refugia for black poplar, one based in Spain and one based in southeastern Europe (Cottrell et al., submitted).
The majority of the clones present in Britain originated from the eastern refugium. The few clones present from the Spanish refugium may have been via human mediated transport.
Hybrid introgression
The P. x euramericana hybrids are represented by a very restricted number of clones in Europe, but these clones are grown in large populations across Europe. In those areas where sexual reproduction still occurs there are anxieties that introgression from these hybrids may further reduce the diversity of the species.
However, work performed by Forest Research (Tabbener & Cottrell, 2003) and other laboratories within EUROPOP clearly demonstrate that the level of introgression is low when male black poplars occur within pollinating distance of female trees. In circumstances where there are no male black poplars the level of introgression from hybrid males in high.
These results have implications for the conservation strategies recommended for the species. A range of molecular markers are now available for the detection of introgression.
What’s of interest
Conservation of black poplar [Archive publication.]
Conservation of black poplar: insights from a DNA fingerprinting approach
Related pages
Useful sites
Genetic conservation molecular studies of oak
 Forest Research has participated in two EU funded studies on the molecular genetics of oak:
Forest Research has participated in two EU funded studies on the molecular genetics of oak:
- FAIROAK – Synthetic maps of gene diversity and provenance performance for utilisation and conservation of oak genetic resources
- OAKFLOW – Intra and interspecific gene flow in oaks.
Morphological differentiation of Quercus robur and Quercus petraea
In the course of the FAIROAK project a method was established to differentiate between the two oak species based on principal component analysis of 13 leaf morphology characters (Kremer et al., 2002).
Structure of genetic diversity
The two British oakwoods which were studied, Roudsea and Dalkeith showed high levels of diversity when studied using microsatellite markers. The diversity was structured across relatively short distances so that trees growing in close proximity tended to show greater similarity to each other than those growing further apart (Cottrell et al., 2002).
Glacial origins
Collaborative work during FAIROAK sampled 2600 oak populations across Europe in order to determine the number of glacial refugia and the postglacial routes of colonisation of oaks across Europe. The results of this enormous effort have been published in a Special Issue of Molecular Ecology (Volume 156).
The results reveal that Britain and Ireland were entirely colonised by material originating from a Spanish refugium which migrated northwards through France and into Britain. The material which entered Britain contained three Iberian haplotypes which showed a distinct distribution pattern, one of which is largely western, another which is central and a third which is largely eastern.
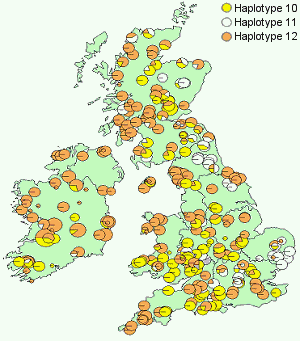
cpDNA variation in British oak
Related pages
Useful sites
Genetic conservation molecular studies of wild cherry
Reproduction

 Wild cherry is a diploid member of the Rosaceae and is a frequent component of woodland margins throughout Europe. It reproduces both sexually (insect pollinated, bird and mammal seed dispersal) and asexually via suckering.
Wild cherry is a diploid member of the Rosaceae and is a frequent component of woodland margins throughout Europe. It reproduces both sexually (insect pollinated, bird and mammal seed dispersal) and asexually via suckering.
A DEFRA/Forestry Commission funded project explored the clonal structure of cherry in two semi-natural, ancient woodlands in Kent and found that 55% of the trees were derived from asexual reproduction.
Clonal groups
The largest clonal patch consisted of 65 trees. Clonal groups were generally non-exclusive in their distribution and displayed a ‘guerrilla’ strategy where further single and multi-ramet genotypes commonly shared the territory occupied by the dominant clone. Evidence of somatic mutation within genets was also observed in both populations.
Recruitment
Recruitment, both asexual and sexual, was markedly higher in the years following major winter storms and attendant disturbance of the canopy. High spatial genetic structure (Sp 0.030-0.045) was observed in the sexually derived trees and kinship coefficients were significant up to 120 m.
Pollen
Pollen flow was also examined using 13 polymorphic microsatellite loci and 54% of paternity was based on pollen from within the two woodlands. A young plantation adjacent to the semi-natural population contributed less than 7% to the paternity of the seeds. Progeny kinship (common paternity) was highest where a mother tree was surrounded by many ramets of a single, compatible clone consisting of sizeable, adult trees. Due to differences in compatibility, close neighbours could be pollinated by very different components of the pollen cloud.
Using a real-time polymerase chain reaction (PCR) approach to monitor black grouse populations
Conservation activities
 The black grouse (Tetrao tetrix) is a large game bird closely related to the capercaillie. Although widespread in Britain until the latter half of the 19th century, it is now one of the most rapidly declining bird species in the UK with a distribution confined to some upland areas of Scotland, Wales and the North of England.
The black grouse (Tetrao tetrix) is a large game bird closely related to the capercaillie. Although widespread in Britain until the latter half of the 19th century, it is now one of the most rapidly declining bird species in the UK with a distribution confined to some upland areas of Scotland, Wales and the North of England.
Black grouse has now been identified as a species of high conservation concern in the UK and conservation activities have centred on improving black grouse habitat.
This has involved activities such as thinning trees in commercial plantations to create more open ground and patchy forest edges, as well as reducing herbivore impacts on moorland edges to encourage black grouse. Attention has also focussed on understanding whether techniques such as heather burning and cutting can benefit this species.
Using a real-time PCR assay
Monitoring the success of these conservation activities is difficult because black grouse are both rare and shy.
To date, population monitoring relies on counting males during the display period when they are at their most visible. However, this is time consuming and the number of actively displaying males can vary with time of day and weather. An alternative approach is to count the faecal droppings of black grouse, but this has proven somewhat problematic as it is sometimes difficult to distinguish the droppings of black grouse from other tetraonid bird species.
To resolve this difficulty we developed a real-time PCR based assay to distinguish between faecal droppings from black grouse and two other tetraonid species that can occur sympatrically; red grouse (Lagopus lagopus scotica) and capercaillie (Tetrao urogallus). The real-time PCR approach is several orders of magnitude more sensitive than traditional PCR methods, with gel-based detection and is, therefore, appropriate for field collected faecal samples which tend to yield poor quality DNA in low copy number.
We have also designed our assay based upon variation in mitochondrial DNA, which maximises the potential for successful identification because of a larger copy number of mitochondrial DNA molecules per cell.
 Black poplar is Britain’s rarest native tree and it is estimated that only about 7000 individual trees remain of this once common tree in Britain. It has become endangered because its natural habitat of flooded winter meadows has been largely lost due to drainage of agricultural land, canalisation of rivers and urbanisation. As a result of this the British population is unable to regenerate sexually and many of the existing trees are ancient and nearing the end of their natural lives. There has been little vegetative propagation of the species in Britain since the introduction of the faster growing P. x euramericana hybrids in the nineteenth century.
Black poplar is Britain’s rarest native tree and it is estimated that only about 7000 individual trees remain of this once common tree in Britain. It has become endangered because its natural habitat of flooded winter meadows has been largely lost due to drainage of agricultural land, canalisation of rivers and urbanisation. As a result of this the British population is unable to regenerate sexually and many of the existing trees are ancient and nearing the end of their natural lives. There has been little vegetative propagation of the species in Britain since the introduction of the faster growing P. x euramericana hybrids in the nineteenth century. Forest Research has participated in two
Forest Research has participated in two 

 Wild cherry is a diploid member of the Rosaceae and is a frequent component of woodland margins throughout Europe. It reproduces both sexually (insect pollinated, bird and mammal seed dispersal) and asexually via suckering.
Wild cherry is a diploid member of the Rosaceae and is a frequent component of woodland margins throughout Europe. It reproduces both sexually (insect pollinated, bird and mammal seed dispersal) and asexually via suckering. The black grouse (Tetrao tetrix) is a large game bird closely related to the capercaillie. Although widespread in Britain until the latter half of the 19th century, it is now one of the most rapidly declining bird species in the UK with a distribution confined to some upland areas of Scotland, Wales and the North of England.
The black grouse (Tetrao tetrix) is a large game bird closely related to the capercaillie. Although widespread in Britain until the latter half of the 19th century, it is now one of the most rapidly declining bird species in the UK with a distribution confined to some upland areas of Scotland, Wales and the North of England.